Geographic detectors(geodetector) Model
Wenbo Lv
Source:vignettes/Geographic-detector.Rmd
Geographic-detector.RmdThis vignette explains how to run a geodetector model in
spEcula package.
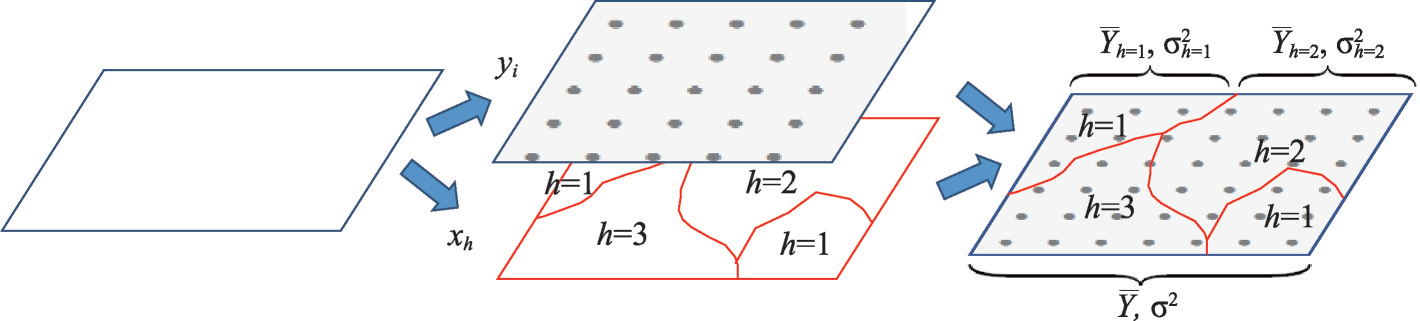
Schematic diagram of Geographic detectors(geodetector) model
basical geographic detector
Load package and pre-processing data.
See layers in NTDs.gpkg:
ntdspath = system.file("extdata", "NTDs.gpkg",package = 'spEcula')
st_layers(ntdspath)
## Driver: GPKG
## Available layers:
## layer_name geometry_type features fields crs_name
## 1 disease Polygon 189 2 unknown
## 2 watershed Polygon 9 2 unknown
## 3 elevation Multi Polygon 7 2 unknown
## 4 soiltype Multi Polygon 6 2 unknownIn NTDs.gpkg, disease is a dependent variable, which is
a continuous numerical variable, while others are independent and
discrete variables.
Now we need to put these layers together:
watershed = read_sf(ntdspath,layer = 'watershed')
elevation = read_sf(ntdspath,layer = 'elevation')
soiltype = read_sf(ntdspath,layer = 'soiltype')
disease = read_sf(ntdspath,layer = 'disease')Plot them together:
library(cowplot)
f1 = ggplot(data = disease) +
geom_sf(aes(fill = incidence),lwd = .1,color = 'grey') +
viridis::scale_fill_viridis(option="mako", direction = -1) +
theme_bw() +
theme(
axis.text = element_blank(),
axis.ticks = element_blank(),
axis.title = element_blank(),
panel.grid = element_blank(),
legend.position = 'inside',
legend.position.inside = c(.1,.25),
legend.background = element_rect(fill = 'transparent',color = NA)
)
f2 = ggplot(data = watershed) +
geom_sf(aes(fill = watershed),lwd = .1,color = 'grey') +
tidyterra::scale_fill_whitebox_c() +
coord_sf(crs = NULL) +
theme_bw() +
theme(
axis.text = element_blank(),
axis.ticks = element_blank(),
axis.title = element_blank(),
panel.grid = element_blank(),
legend.position = 'inside',
legend.position.inside = c(.1,.25),
legend.background = element_rect(fill = 'transparent',color = NA)
)
f3 = ggplot(data = elevation) +
geom_sf(aes(fill = elevation),lwd = .1,color = 'grey') +
tidyterra::scale_fill_hypso_c() +
theme_bw() +
theme(
axis.text = element_blank(),
axis.ticks = element_blank(),
axis.title = element_blank(),
panel.grid = element_blank(),
legend.position = 'inside',
legend.position.inside = c(.1,.25),
legend.background = element_rect(fill = 'transparent',color = NA)
)
f4 = ggplot(data = soiltype) +
geom_sf(aes(fill = soiltype),lwd = .1,color = 'grey') +
tidyterra::scale_fill_wiki_c() +
theme_bw() +
theme(
axis.text = element_blank(),
axis.ticks = element_blank(),
axis.title = element_blank(),
panel.grid = element_blank(),
legend.position = 'inside',
legend.position.inside = c(.1,.25),
legend.background = element_rect(fill = 'transparent',color = NA)
)
plot_grid(f1,f2,f3,f4, nrow = 2,label_fontfamily = 'serif',
labels = paste0('(',letters[1:4],')'),
label_fontface = 'plain',label_size = 10,
hjust = -1.5,align = 'hv') -> p
p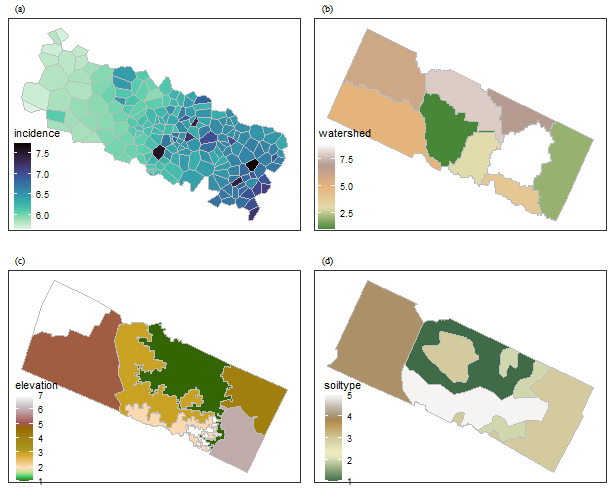
Attribute spatial join
NTDs = disease %>%
st_centroid() %>%
st_join(watershed[,"watershed"]) %>%
st_join(elevation[,"elevation"]) %>%
st_join(soiltype[,"soiltype"])Check whether has NA in NTDs:
NTDs %>%
dplyr::filter(if_any(everything(),~is.na(.x)))
## Simple feature collection with 4 features and 5 fields
## Geometry type: POINT
## Dimension: XY
## Bounding box: xmin: 301567.5 ymin: 3989433 xmax: 318763.3 ymax: 3991906
## Projected CRS: +proj=aea +lat_0=0 +lon_0=105 +lat_1=25 +lat_2=47 +x_0=0 +y_0=0 +datum=WGS84 +units=m +no_defs
## # A tibble: 4 × 6
## SP_ID incidence geom watershed elevation soiltype
## * <chr> <dbl> <POINT [m]> <int> <int> <int>
## 1 141 6.48 (318763.3 3991847) 9 NA 3
## 2 165 6.53 (316574.1 3989433) 4 NA 2
## 3 166 6.43 (311439.1 3990674) 4 NA 2
## 4 188 6.26 (301567.5 3991906) NA 2 3
NTDs %>%
dplyr::filter(if_all(everything(),~!is.na(.x))) -> NTDsFactor detector
NTDs = st_drop_geometry(NTDs)
fd = ssh.test(incidence ~ watershed + elevation + soiltype,
data = NTDs,type = 'factor')
fd
## Spatial Stratified Heterogeneity Test
##
## Factor detector| variable | Q-statistic | P-value |
|---|---|---|
| watershed | 0.6378 | 0.0001288 |
| elevation | 0.6067 | 0.04338 |
| soiltype | 0.3857 | 0.3721 |
Interaction detector
id = ssh.test(incidence ~ watershed + elevation + soiltype,
data = NTDs,type = 'interaction')
id
## Spatial Stratified Heterogeneity Test
##
## Interaction detector| Interactive variable | Interaction |
|---|---|
| watershed ∩ elevation | Enhance, bi- |
| watershed ∩ soiltype | Enhance, bi- |
| elevation ∩ soiltype | Enhance, bi- |
Risk detector
rd = ssh.test(incidence ~ watershed + elevation + soiltype,
data = NTDs,type = 'risk')
rd
## Spatial Stratified Heterogeneity Test
##
## Risk detector
##
## --------------------------------------
## Variable elevation:
##
## |1 |2 |3 |4 |5 |6 |7 |
## |:---|:---|:---|:---|:---|:---|:---|
## |NA |Yes |Yes |Yes |Yes |Yes |Yes |
## |Yes |NA |No |Yes |Yes |Yes |Yes |
## |Yes |No |NA |Yes |Yes |Yes |Yes |
## |Yes |Yes |Yes |NA |Yes |Yes |Yes |
## |Yes |Yes |Yes |Yes |NA |Yes |Yes |
## |Yes |Yes |Yes |Yes |Yes |NA |Yes |
## |Yes |Yes |Yes |Yes |Yes |Yes |NA |
## --------------------------------------
## Variable soiltype:
##
## |1 |2 |3 |4 |5 |
## |:---|:---|:---|:---|:---|
## |NA |Yes |Yes |Yes |No |
## |Yes |NA |No |Yes |Yes |
## |Yes |No |NA |Yes |Yes |
## |Yes |Yes |Yes |NA |Yes |
## |No |Yes |Yes |Yes |NA |
## --------------------------------------
## Variable watershed:
##
## |1 |2 |3 |4 |5 |6 |7 |8 |9 |
## |:---|:---|:---|:---|:---|:---|:---|:---|:---|
## |NA |Yes |Yes |Yes |Yes |Yes |Yes |Yes |Yes |
## |Yes |NA |Yes |No |Yes |Yes |Yes |Yes |Yes |
## |Yes |Yes |NA |Yes |Yes |Yes |No |No |No |
## |Yes |No |Yes |NA |Yes |Yes |Yes |Yes |Yes |
## |Yes |Yes |Yes |Yes |NA |No |Yes |Yes |Yes |
## |Yes |Yes |Yes |Yes |No |NA |Yes |Yes |Yes |
## |Yes |Yes |No |Yes |Yes |Yes |NA |No |No |
## |Yes |Yes |No |Yes |Yes |Yes |No |NA |Yes |
## |Yes |Yes |No |Yes |Yes |Yes |No |Yes |NA |You can change the significant interval by assign alpha
argument,the default value of alpha argument is
0.95.
rd99 = ssh.test(incidence ~ watershed + elevation + soiltype,
data = NTDs,type = 'risk',alpha = 0.99)
rd99
## Spatial Stratified Heterogeneity Test
##
## Risk detector
##
## --------------------------------------
## Variable elevation:
##
## |1 |2 |3 |4 |5 |6 |7 |
## |:---|:---|:---|:---|:---|:---|:---|
## |NA |Yes |Yes |Yes |Yes |Yes |Yes |
## |Yes |NA |No |Yes |Yes |Yes |Yes |
## |Yes |No |NA |Yes |Yes |Yes |Yes |
## |Yes |Yes |Yes |NA |Yes |Yes |Yes |
## |Yes |Yes |Yes |Yes |NA |Yes |Yes |
## |Yes |Yes |Yes |Yes |Yes |NA |Yes |
## |Yes |Yes |Yes |Yes |Yes |Yes |NA |
## --------------------------------------
## Variable soiltype:
##
## |1 |2 |3 |4 |5 |
## |:---|:---|:---|:---|:---|
## |NA |Yes |Yes |Yes |No |
## |Yes |NA |No |Yes |Yes |
## |Yes |No |NA |Yes |Yes |
## |Yes |Yes |Yes |NA |Yes |
## |No |Yes |Yes |Yes |NA |
## --------------------------------------
## Variable watershed:
##
## |1 |2 |3 |4 |5 |6 |7 |8 |9 |
## |:---|:---|:---|:---|:---|:---|:---|:---|:---|
## |NA |Yes |Yes |Yes |Yes |Yes |Yes |Yes |Yes |
## |Yes |NA |Yes |No |Yes |Yes |Yes |Yes |Yes |
## |Yes |Yes |NA |Yes |Yes |Yes |No |No |No |
## |Yes |No |Yes |NA |Yes |Yes |Yes |Yes |Yes |
## |Yes |Yes |Yes |Yes |NA |No |Yes |Yes |Yes |
## |Yes |Yes |Yes |Yes |No |NA |Yes |Yes |Yes |
## |Yes |Yes |No |Yes |Yes |Yes |NA |No |No |
## |Yes |Yes |No |Yes |Yes |Yes |No |NA |Yes |
## |Yes |Yes |No |Yes |Yes |Yes |No |Yes |NA |Ecological detector
ed = ssh.test(incidence ~ watershed + elevation + soiltype,
data = NTDs,type = 'ecological')
ed
## Spatial Stratified Heterogeneity Test
##
## ecological detector
##
## --------------------------------------
##
##
## | |elevation |soiltype |
## |:---------|:---------|:--------|
## |watershed |No |No |
## |elevation |NA |No |You can also change the significant interval by assign
alpha argument,the default value of alpha
argument is 0.95.
ed99 = ssh.test(incidence ~ watershed + elevation + soiltype,
data = NTDs,type = 'ecological',alpha = 0.99)
ed99
## Spatial Stratified Heterogeneity Test
##
## ecological detector
##
## --------------------------------------
##
##
## | |elevation |soiltype |
## |:---------|:---------|:--------|
## |watershed |No |No |
## |elevation |NA |No |optimal parameters geographic detector(OPGD)
library(terra)
fvcpath = "https://github.com/SpatLyu/rdevdata/raw/main/FVC.tif"
fvc = terra::rast(paste0("/vsicurl/",fvcpath))
fvc
## class : SpatRaster
## dimensions : 418, 568, 13 (nrow, ncol, nlyr)
## resolution : 1000, 1000 (x, y)
## extent : -92742.16, 475257.8, 3591385, 4009385 (xmin, xmax, ymin, ymax)
## coord. ref. : Asia_North_Albers_Equal_Area_Conic
## source : FVC.tif
## names : fvc, premax, premin, presum, tmpmax, tmpmin, ...
## min values : 0.1363270, 109.8619, 2.00000, 3783.904, 9.289694, -11.971293, ...
## max values : 0.9596695, 249.9284, 82.74928, 8549.112, 26.781870, 1.322163, ...
names(fvc)
## [1] "fvc" "premax" "premin" "presum" "tmpmax" "tmpmin" "tmpavg" "pop" "ntl"
## [10] "lulc" "elev" "slope" "aspect"Convert data from SpatRaster to
tibble
fvc = as_tibble(terra::as.data.frame(fvc,na.rm = T))
head(fvc)
## # A tibble: 6 × 13
## fvc premax premin presum tmpmax tmpmin tmpavg pop ntl lulc elev slope aspect
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 0.198 163. 7.95 3956. 20.8 -7.53 8.05 1.90 6.60 10 1758. 2.65 176.
## 2 0.193 161. 6.80 3892. 20.7 -7.55 8.02 1.20 4.91 10 1754. 3.45 170.
## 3 0.192 160. 5.24 3842. 20.9 -7.48 8.15 0.547 3.75 10 1722. 3.96 139.
## 4 0.189 159. 5 3808. 21.1 -7.39 8.35 0.542 3.99 10 1672. 2.90 111.
## 5 0.208 164. 9.98 4051. 20.6 -7.59 7.97 10.4 7.10 10 1780. 1.94 99.5
## 6 0.196 163. 8.15 3973. 20.7 -7.53 8.03 9.31 6.56 10 1755. 3.01 99.6Determine optimal discretization parameters
Only lulc is a discrete category variable in the
fvc data, we need to discretize others. We can use
gd_bestunidisc to discretize them based on geodetector
q-statistic.
tictoc::tic()
g = gd_bestunidisc(fvc ~ .,data = select(fvc,-lulc),discnum = 2:15,cores = 6)
tictoc::toc()
## 18.26 sec elapsed
g
## $x
## [1] "aspect" "elev" "ntl" "pop" "premax" "premin" "presum" "slope" "tmpavg"
## [10] "tmpmax" "tmpmin"
##
## $k
## [1] 15 15 15 15 14 15 15 13 15 15 15
##
## $method
## [1] "equal" "quantile" "fisher" "quantile" "fisher" "fisher" "fisher"
## [8] "fisher" "quantile" "quantile" "fisher"
##
## $disv
## # A tibble: 136,243 × 11
## aspect elev ntl pop premax premin presum slope tmpavg tmpmax tmpmin
## <int> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int>
## 1 8 12 3 1 7 1 1 4 4 8 2
## 2 8 12 2 1 6 1 1 5 4 8 2
## 3 6 12 2 1 6 1 1 6 4 9 2
## 4 5 12 2 1 6 1 1 4 5 10 2
## 5 5 13 3 4 7 2 1 3 4 8 2
## 6 5 12 3 3 7 2 1 5 4 8 2
## 7 6 12 2 1 6 1 1 6 4 9 3
## 8 7 12 2 1 6 1 1 5 5 10 3
## 9 8 12 2 1 6 1 1 3 5 10 3
## 10 7 12 3 1 6 1 1 5 5 10 3
## # ℹ 136,233 more rows
new.fvc = g$disv
new.fvc
## # A tibble: 136,243 × 11
## aspect elev ntl pop premax premin presum slope tmpavg tmpmax tmpmin
## <int> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int>
## 1 8 12 3 1 7 1 1 4 4 8 2
## 2 8 12 2 1 6 1 1 5 4 8 2
## 3 6 12 2 1 6 1 1 6 4 9 2
## 4 5 12 2 1 6 1 1 4 5 10 2
## 5 5 13 3 4 7 2 1 3 4 8 2
## 6 5 12 3 3 7 2 1 5 4 8 2
## 7 6 12 2 1 6 1 1 6 4 9 3
## 8 7 12 2 1 6 1 1 5 5 10 3
## 9 8 12 2 1 6 1 1 3 5 10 3
## 10 7 12 3 1 6 1 1 5 5 10 3
## # ℹ 136,233 more rowsThe new.fvc is the discrete result of the optimal
discretization parameter based on the Q statistic of the geographic
detector,we can combine it with fvc and lulc
col in fvc tibble now.
new.fvc = bind_cols(select(fvc,fvc,lulc),new.fvc)
new.fvc
## # A tibble: 136,243 × 13
## fvc lulc aspect elev ntl pop premax premin presum slope tmpavg tmpmax tmpmin
## <dbl> <dbl> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int>
## 1 0.198 10 8 12 3 1 7 1 1 4 4 8 2
## 2 0.193 10 8 12 2 1 6 1 1 5 4 8 2
## 3 0.192 10 6 12 2 1 6 1 1 6 4 9 2
## 4 0.189 10 5 12 2 1 6 1 1 4 5 10 2
## 5 0.208 10 5 13 3 4 7 2 1 3 4 8 2
## 6 0.196 10 5 12 3 3 7 2 1 5 4 8 2
## 7 0.191 10 6 12 2 1 6 1 1 6 4 9 3
## 8 0.185 10 7 12 2 1 6 1 1 5 5 10 3
## 9 0.174 10 8 12 2 1 6 1 1 3 5 10 3
## 10 0.166 10 7 12 3 1 6 1 1 5 5 10 3
## # ℹ 136,233 more rowsRun geodetector
Then ,we can run geodetector model by ssh.test()
function.
ssh.test(fvc ~ .,data = new.fvc,type = 'factor')
## Spatial Stratified Heterogeneity Test
##
## Factor detector| variable | Q-statistic | P-value |
|---|---|---|
| presum | 0.6406 | 6.684e-10 |
| lulc | 0.5533 | 9.106e-10 |
| premin | 0.4427 | 4.262e-10 |
| tmpmin | 0.4068 | 3.583e-10 |
| tmpmax | 0.2284 | 5.111e-10 |
| elev | 0.209 | 1.5e-10 |
| tmpavg | 0.197 | 6.833e-10 |
| slope | 0.1911 | 7.183e-10 |
| pop | 0.1856 | 3.221e-10 |
| premax | 0.1323 | 4.211e-10 |
| ntl | 0.02156 | 5.865e-10 |
| aspect | 0.00741 | 5.448e-10 |
ssh.test(fvc ~ .,data = new.fvc,type = 'interaction')
## Spatial Stratified Heterogeneity Test
##
## Interaction detector| Interactive variable | Interaction |
|---|---|
| lulc ∩ aspect | Enhance, nonlinear |
| lulc ∩ elev | Enhance, bi- |
| lulc ∩ ntl | Enhance, nonlinear |
| lulc ∩ pop | Enhance, bi- |
| lulc ∩ premax | Enhance, bi- |
| lulc ∩ premin | Enhance, bi- |
| lulc ∩ presum | Enhance, bi- |
| lulc ∩ slope | Enhance, bi- |
| lulc ∩ tmpavg | Enhance, bi- |
| lulc ∩ tmpmax | Enhance, bi- |
| lulc ∩ tmpmin | Enhance, bi- |
| aspect ∩ elev | Enhance, nonlinear |
| aspect ∩ ntl | Enhance, nonlinear |
| aspect ∩ pop | Enhance, nonlinear |
| aspect ∩ premax | Enhance, nonlinear |
| aspect ∩ premin | Enhance, nonlinear |
| aspect ∩ presum | Weaken, uni- |
| aspect ∩ slope | Enhance, nonlinear |
| aspect ∩ tmpavg | Enhance, nonlinear |
| aspect ∩ tmpmax | Enhance, nonlinear |
| aspect ∩ tmpmin | Enhance, nonlinear |
| elev ∩ ntl | Enhance, nonlinear |
| elev ∩ pop | Enhance, bi- |
| elev ∩ premax | Enhance, nonlinear |
| elev ∩ premin | Enhance, bi- |
| elev ∩ presum | Enhance, bi- |
| elev ∩ slope | Enhance, bi- |
| elev ∩ tmpavg | Enhance, bi- |
| elev ∩ tmpmax | Enhance, nonlinear |
| elev ∩ tmpmin | Enhance, bi- |
| ntl ∩ pop | Enhance, nonlinear |
| ntl ∩ premax | Enhance, nonlinear |
| ntl ∩ premin | Enhance, nonlinear |
| ntl ∩ presum | Enhance, nonlinear |
| ntl ∩ slope | Enhance, nonlinear |
| ntl ∩ tmpavg | Enhance, nonlinear |
| ntl ∩ tmpmax | Enhance, nonlinear |
| ntl ∩ tmpmin | Enhance, nonlinear |
| pop ∩ premax | Enhance, nonlinear |
| pop ∩ premin | Enhance, bi- |
| pop ∩ presum | Enhance, bi- |
| pop ∩ slope | Enhance, bi- |
| pop ∩ tmpavg | Enhance, nonlinear |
| pop ∩ tmpmax | Enhance, nonlinear |
| pop ∩ tmpmin | Enhance, bi- |
| premax ∩ premin | Enhance, nonlinear |
| premax ∩ presum | Enhance, bi- |
| premax ∩ slope | Enhance, nonlinear |
| premax ∩ tmpavg | Enhance, nonlinear |
| premax ∩ tmpmax | Enhance, nonlinear |
| premax ∩ tmpmin | Enhance, nonlinear |
| premin ∩ presum | Enhance, bi- |
| premin ∩ slope | Enhance, bi- |
| premin ∩ tmpavg | Enhance, bi- |
| premin ∩ tmpmax | Enhance, bi- |
| premin ∩ tmpmin | Enhance, bi- |
| presum ∩ slope | Enhance, bi- |
| presum ∩ tmpavg | Enhance, bi- |
| presum ∩ tmpmax | Enhance, bi- |
| presum ∩ tmpmin | Enhance, bi- |
| slope ∩ tmpavg | Enhance, bi- |
| slope ∩ tmpmax | Enhance, bi- |
| slope ∩ tmpmin | Enhance, bi- |
| tmpavg ∩ tmpmax | Enhance, nonlinear |
| tmpavg ∩ tmpmin | Enhance, nonlinear |
| tmpmax ∩ tmpmin | Enhance, nonlinear |